Importing raw (unprocessed) Affymetrix microarray data
Overview
Teaching: 10 min
Exercises: 5 minLearning Objectives
Be able to obtain supplemental data
Be able to explain and use the differences between GEO data types.
Understand the concept of the ExpressionSet class of objects.
We now have processed data for two series, GSE33146 and [GSE66417][GEE66417]. We need to get the unprocessed data to understand the processing.
Getting the raw data for the series using GEOquery
Getting the raw data (CEL files for Affymetrix data) is distinct from getting the processed ata. The CEL files can be pretty large, and sometimes the download can fail, even with a good connection.
Nonetheless, if you want to do it, getGEOSuppFiles() will download all the
supplementary files for the GEO accession. Note that it doesn’t process or even parse the files,
since there are many different types of supplementary files on GEO and R doesn’t know the format ahead of time. It does, however, return the file paths.
## If you have an awesome connection and a lot of time
filePaths <- rownames(getGEOSuppFiles('GSE33146'))
Using locally cached version of supplementary file(s) GSE33146 found here:
/home/gtk/work/Microarrays-R/_episodes_rmd/GSE33146/GSE33146_RAW.tar
untar(filePaths,exdir="GSE33146")
filePaths <- rownames(getGEOSuppFiles('GSE66417'))
Using locally cached version of supplementary file(s) GSE66417 found here:
/home/gtk/work/Microarrays-R/_episodes_rmd/GSE66417/GSE66417_RAW.tar
untar(filePaths,exdir="GSE66417")
In the interest of time, don’t do that. Instead, use the files you have downloaded from Canvas and unpacked into a directory named for each series. You can leave them compressed.
Reading CEL data using the oligo package.
The oligo package provides functions for handling Affymetrix data,
including CEL file data. The function oligo::read.celfiles() does
the work. This function takes in a vector of filenames as the
argument. We can manually type the file names into a vector, using the
c() function. Alternatively, we can use list.celfiles() command
provided by oligoClasses, which is installed when we install oligo. list.celfiles() will list all the files
ending with the .cel extension (CEL files) in a directory, (by
default the current working directory). Helpfully:
- the argument
listGzipped=TRUEwill find compressed CEL files. - the argument
full.names=TRUEwill include the directory name, so the results can be passed toread.celfiles().
Therefore, the following needs to be done:
- Use
oligoClasseslist.celfiles(GSE33146)with the appropriate arguments to generate a vector containing the name of all the CEL files. - Use
read.celfiles()to read in the CEL files.
Try it!
Try to read the CEL files for both data sets into R using the information provided above.
Solution
library(oligo) library(oligoClasses) gse33146_celdata <- read.celfiles(list.celfiles('GSE33146',full.names=TRUE,listGzipped=TRUE))Reading in : GSE33146/GSM820817.CEL.gz Reading in : GSE33146/GSM820818.CEL.gz Reading in : GSE33146/GSM820819.CEL.gz Reading in : GSE33146/GSM820820.CEL.gz Reading in : GSE33146/GSM820821.CEL.gz Reading in : GSE33146/GSM820822.CEL.gzgse66417_celdata <- read.celfiles(list.celfiles('GSE66417',full.names=TRUE,listGzipped=TRUE))Reading in : GSE66417/GSM1622170_Jurkat-Ctrl-RD1_HuGene-2_0-st_.CEL.gz Reading in : GSE66417/GSM1622189_Jurkat-Ctrl-RD2_HuGene-2_0-st_.CEL.gz Reading in : GSE66417/GSM1622191_Jurkat-Ctrl-RD3_HuGene-2_0-st_.CEL.gz Reading in : GSE66417/GSM1622194_Jurkat-Ixazomib-RD4_HuGene-2_0-st_.CEL.gz Reading in : GSE66417/GSM1622196_Jurkat-Ixazomib-RD5_HuGene-2_0-st_.CEL.gz Reading in : GSE66417/GSM1622198_Jurkat-Ixazomib-RD6_HuGene-2_0-st_.CEL.gz Reading in : GSE66417/GSM1622200_L540-Ctrl-RD7_HuGene-2_0-st_.CEL.gz Reading in : GSE66417/GSM1622202_L540-Ctrl-RD8_HuGene-2_0-st_.CEL.gz Reading in : GSE66417/GSM1622204_L540-Ctrl-RD9_HuGene-2_0-st_.CEL.gz Reading in : GSE66417/GSM1622206_L540-Ixazomib-RD10_HuGene-2_0-st_.CEL.gz Reading in : GSE66417/GSM1622209_L540-Ixazomib-RD11_HuGene-2_0-st_.CEL.gz Reading in : GSE66417/GSM1622211_L540-Ixazomib-RD12_HuGene-2_0-st_.CEL.gz
Once you have the CEL file data, you can get a pseudo-image of the chip intensities.
image(gse33146_celdata[,1])
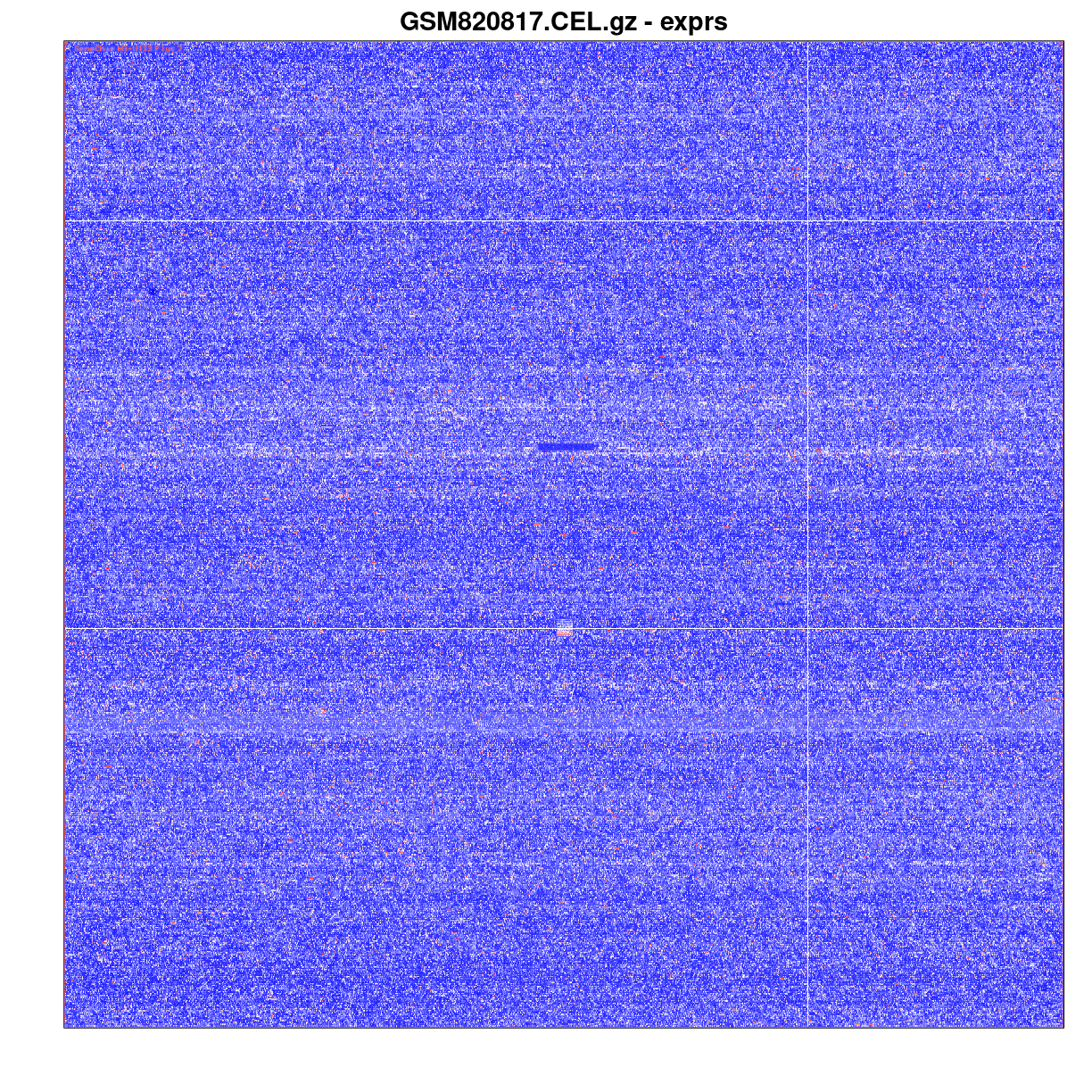
An image of a CEL file
Note that read.celfiles loaded data packages from Bioconductor containing information on the platform design for each chip type.
The risk of using
list.celfiles()Using
list.celfiles()to provide the files toread.celfiles()can be risky.list.celfiles()provides a list of files in lexicographic order. This is probably the same order as the files in your GSE, but can you be sure? Ultimately, you need to use the metadata from the series to ensure that the rows of the phenoData match the columns of the assayData That is the subject of our next episode.
oligooraffy?The
affypackage also has alist.celfiles()function (with a slightly different interface), and offers aread.AffyBatch()function to read celfiles. However, theaffypackage can only work with 3′ biased Affymetrix arrays, sooligois preferred
Key Points
GEO data types have enough similarities to allow data access, but enough differences to require specific type-specific steps.
The ExpressionSet class of object contains slots for different information associated with a microarray experiment.