Motif enrichment/discovery
Overview
Teaching: 15 min
Exercises: 15 minLearning Objectives
Appreciate the utility of motif enrichment/discovery from Chip-Seq data
Complete a motif analysis using the Chip-seq data provided
Comment on the results obtained from the motif analysis
What is motif discovery?
“Motif analysis is useful for much more than just identifying the causal DNA-binding motif in TF ChIP-seq peaks. When the motif of the ChIPed protein is already known, motif analysis provides validation of the success of the experiment. Even when the motif is not known beforehand, identifying a centrally located motif in a large fraction of the peaks by motif analysis is indicative of a successful experiment. Motif analysis can also identify the DNA-binding motifs of other proteins that bind in complex or in conjunction with the ChIPed protein, illuminating the mechanisms of transcriptional regulation. Motif analysis is also useful with histone modification ChIP-seq because it can discover unanticipated sequence signals associated with such marks.”
Excerpt from Bailey T, Krajewski P, Ladunga I, Lefebvre C, Li Q, Liu T, et al. (2013) Practical Guidelines for the Comprehensive Analysis of ChIP-seq Data. PLoS Comput Biol 9(11): e1003326. doi:10.1371/journal.pcbi.1003326
HOMER
First things first, let’s create a folder for HOMER to live in.
cd ~ mkdir HOMER/ cd HOMER/
Now download the file and save it into HOMER/ folder.
And now we run the configureHomer.pl file
perl configureHomer.pl -install
At the end of the installation, configureHomer.pl will ask you to add a line to your .bashrc or .bash_profile file. The line will look something like this:
PATH=$PATH:/path/to/HOMER/.//bin/
Let’s do this!
open your .bashrc or .bash_profile file using your favourite text editor
Put the line in your file:
export PATH=$PATH:/path/to/HOMER/.//bin/save your file and then reload the .bashrc file by typing:
source .bashrc
After we’ve installed HOMER we can run the findMotifsGenome.pl
findMotifsGenome.pl NAME_peaks.bed /path/to/hg19.fa MotifOutput/ -size 200 -mask
And now we can look at the outputs produced by findMotifsGenome.pl by looking in our MotifOutput/ directory. You will notice a bunch of files and two folders homerResults/ and knownResults/. homerResults/ will contain all the files required to generate the homerResults.html and this corresponds to the de novo discovery of motifs. knownResults/ will contain all the files required to generate knownResults.html and will correspond to the enrichment of known motifs.
homerResults.html knownResults.html
The list of discovered motifs will be contained in the file homerMotifs.all.motifs.
Let’s see what’s inside
>WWTTCCCGCC 1-WWTTCCCGCC 5.364976 -1727.061690 0 T:11926.0(46.44%),B:7159.6(29.07%),P:1e-750 Tpos:99.7,Tstd:51.6,Bpos:97.4,Bstd:64.0,StrandBias:0.0,Multiplicity:1.36 0.408 0.140 0.152 0.300 0.292 0.155 0.174 0.379 0.112 0.131 0.221 0.536 0.071 0.211 0.105 0.613 0.100 0.536 0.169 0.195 0.022 0.770 0.176 0.032 0.026 0.736 0.192 0.046 0.046 0.042 0.715 0.197 0.015 0.800 0.173 0.012
Ensemble methods for motif discovery
There are many many motif discovery tools out there. All utilising different algorithms and will have varying outputs. It is best to use a number of tools together and to find the top hits that can be found from those methods.
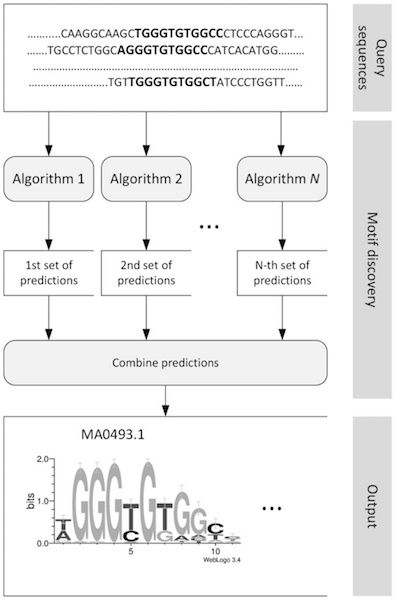
Comparison of results and top result selection: STAMP
A way to find common motifs discovered by your different tools would be to use STAMP.
Here is one I prepared earlier
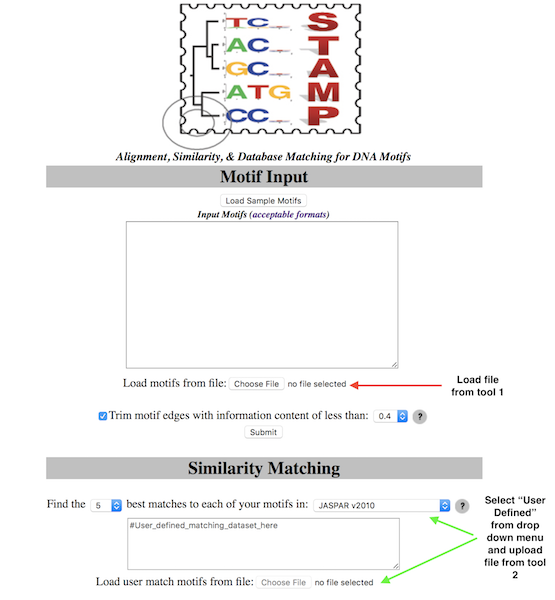
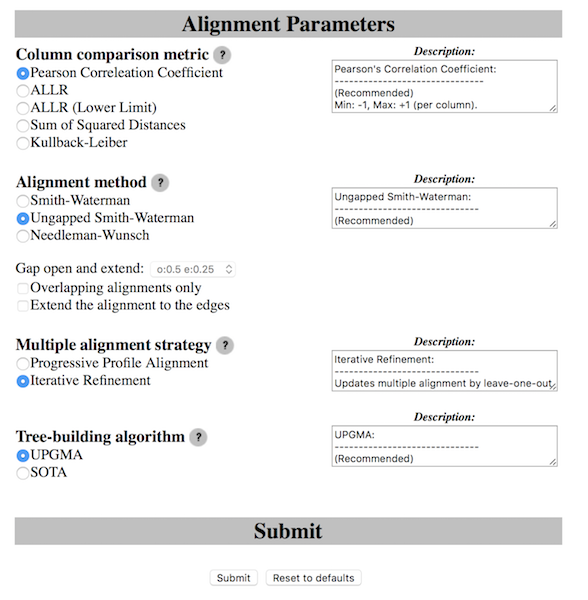
Take note
Unless you really know what your looking for keep the options to default!
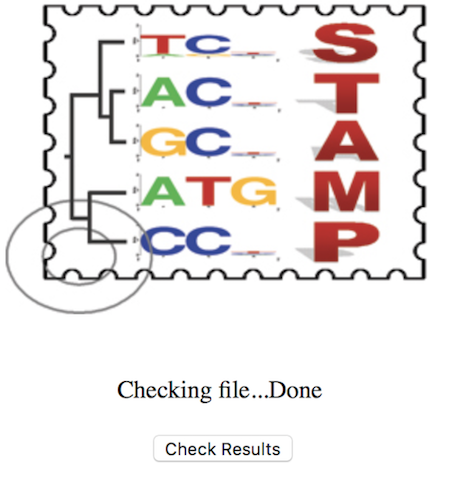
Click the “Check Results” button and it will bring you to a results page. You can then save the html page and download the PDF. Let’s look at the output.
Multiple alignment:

Tree diagram:
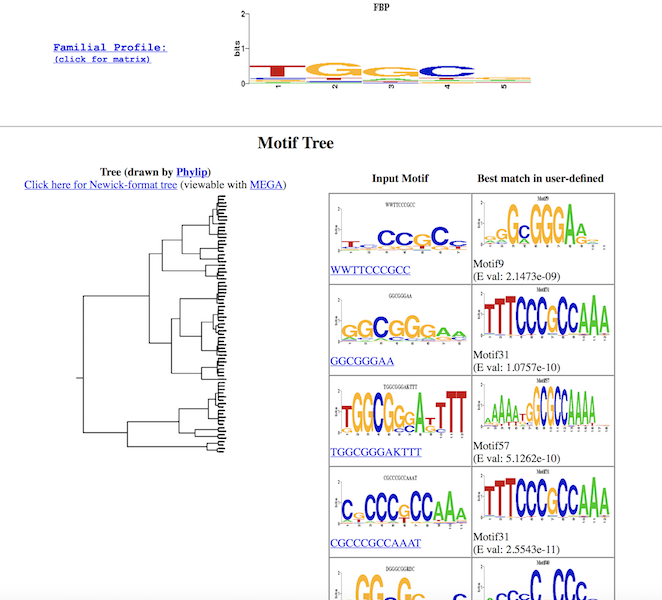
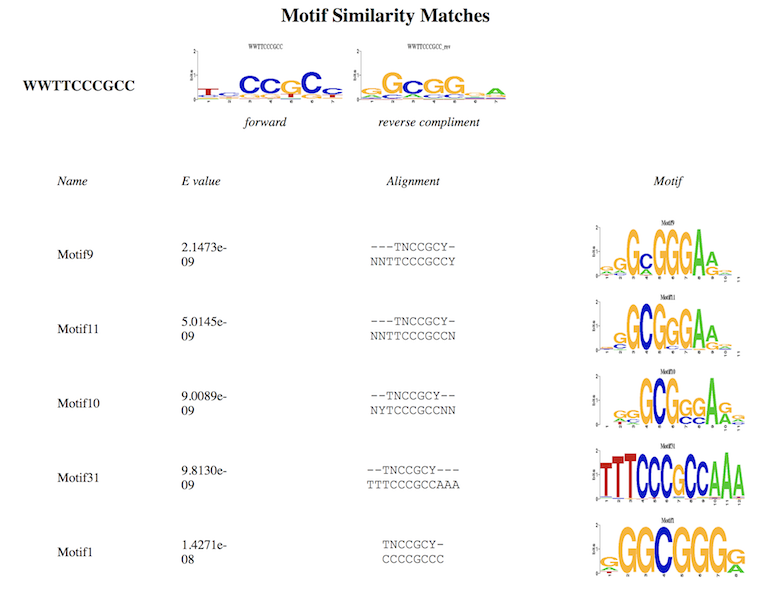
Details of similar motifs from the two datasets:
If your interested to know more about motif discovery
Review of ensemble based motif discovery tools
Review of web based tools for motif discovery
Limitations of De novo motif discovery
Key Points
Motif discovery is not a simple problem and there exist many different techniques to solve the problem.
It is best to take an ensemble approach and to take the results that occur more than once